
Victory Yinka-Banjo is a junior majoring in 6-7: Computer Science and Molecular Biology.

The Department of Electrical Engineering and Computer Science (EECS) is proud to announce multiple promotions.

MIT CSAIL researchers develop advanced machine-learning models that outperform current methods in detecting pancreatic ductal adenocarcinoma.
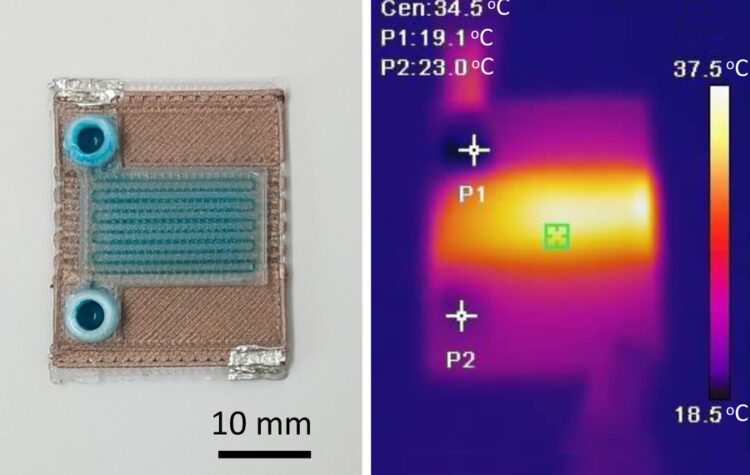
The one-step fabrication process rapidly produces miniature chemical reactors that could be used to detect diseases or analyze substances.

Five MIT faculty, along with seven additional affiliates, are honored for outstanding contributions to medical research.

This ongoing listing of awards and recognitions won by our faculty is added to all year, beginning in September.

Undergraduate engineering and computer science programs are No. 1; undergraduate business program is No. 2.

Department of EECS Assistant Professors Connor Coley and Dylan Hadfield-Menell have been named to the inaugural cohort of AI2050 Early Career Fellows by Schmidt Futures, a philanthropic initiative from Eric and Wendy Schmidt aimed at helping to solve hard problems in AI.

It’s nearing the end of 2021, and we want to celebrate the accomplishments and contributions of our incredible EECS community by sharing some of the awards given by…

Left to right: Álvaro Fernández Galiana, Fatima Hussain, Sirma Orguc, and Rebecca Pinals have been named as Schmidt Science Fellows, an honor created in 2017 to encourage young…